About siVirus
siVirus especially focuses on antiviral siRNA design and provides:
- Highly conserved target sites for designing antiviral siRNAs that would resist viral mutational escapes. siRNA sequences are selected based on their degree of conservation, defined as the proportion of viral sequences that are targeted by the corresponding siRNA, with complete matches (i.e., 21/21 matches). All possible siRNA candidates targeting every other position of user-selected viral sequences are generated and their degrees of conservation are compared. Users can arbitrarily specify a set of viral sequences for the computation; for example, sequences can be selected from a specific geographic region(s) or a specific genotype(s) to design the best siRNAs tailored to specific user needs.
- Effective siRNAs that satisfy efficacy guidelines. We incorporate the guidelines of Ui-Tei et al., Reynolds et al., and Amarzguioui et al. and shows whether each siRNA satisfies these guidelines.
- Off-target minimized siRNAs. siVirus shows the number of off-target hits within two mismatches against human genes. For details, links to the siDirect off-target search pages are also provided. It is desirable to select an siRNA that has less off-target hits.
Examples
Designing siRNAs for HIV-1
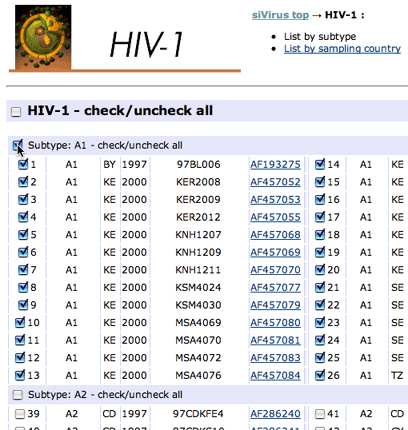
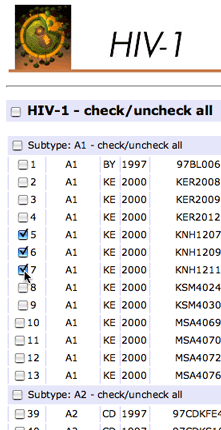
- Select viral sequences for the conservation analysis. For example, select all subtype A1 sequences if you want to design optimal siRNAs for subtype A1. Simply checking all sequences is a good choice if you don't care about subtypes or geographic regions.
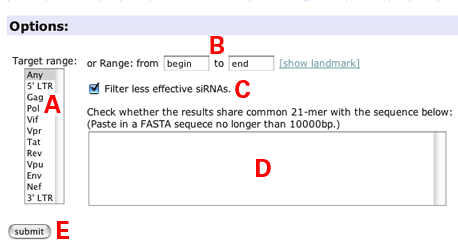
- Options:
- Select gene(s) or genomic region(s) for siRNA design.
- Specify target region by nucleotide position.
- Check here to hide less effective siRNAs (default).
- (not yet implemented)
- Submit button.
- Click submit button to design siRNAs.
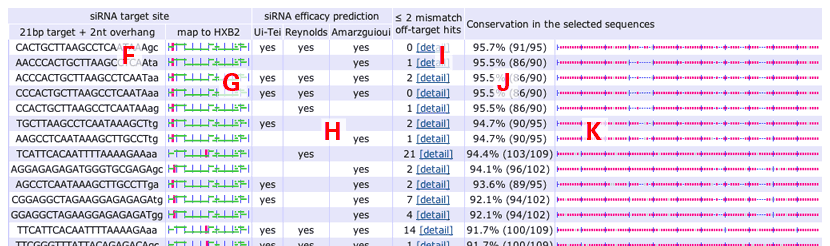
- Results:
- siRNA sequence.
- siRNA position.
- siRNA efficacy predictions.
- Off-target search results. The number of human off-targets within two mismatches.
- Degrees of conservation.
- Conservation in the user-selected sequences.
References
siVirus web server
- If you use this site please acknowledge by citing us:
Naito Y, Ui-Tei K, Nishikawa T, Takebe Y, Saigo K. (2006)
siVirus: web-based antiviral siRNA design software for highly divergent viral sequences.
Nucleic Acids Res., 34, W448-W450.
Viral sequence databases
Designing effective siRNA
- Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. (2004)
Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference.
Nucleic Acids Res., 32, 936-948.
[FullText]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. (2004)
Rational siRNA design for RNA interference.
Nat. Biotechnol., 22, 326-330.
[PubMed]
- Amarzguioui M, Prydz H. (2004)
An algorithm for selection of functional siRNA sequences.
Biochem. Biophys. Res. Commun., 316, 1050-1058.
[PubMed]
Efficient off-target searches for siRNA
- Yamada T, Morishita S. (2005)
Accelerated off-target search algorithm for siRNA.
Bioinformatics, 21, 1316-1324.
[PubMed]
- siDirect web server
Naito Y*, Yamada T*, Ui-Tei K, Morishita S, Saigo K. (2004)
siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference.
Nucleic Acids Res., 32, W124-W129. *Joint First Authors.
[FullText]
Credits
Yuki Naito, Kumiko Ui-Tei, Toru Nishikawa, Yutaka Takebe and Kaoru Saigo used the expertise of biology and informatics, and designed the system. Yuki Naito and Toru Nishikawa together implemented the entire system of siVirus, and developed novel, efficient algorithms for designing antiviral siRNAs. This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan to K.S., K.U.-T. and Y.T., and by grants from the Ministry of Health, Labour and Welfare of Japan to Y.T.
Contact Information
E-mail: siVirus @ RNAi.jp
Last modified on Aug 20, 2006